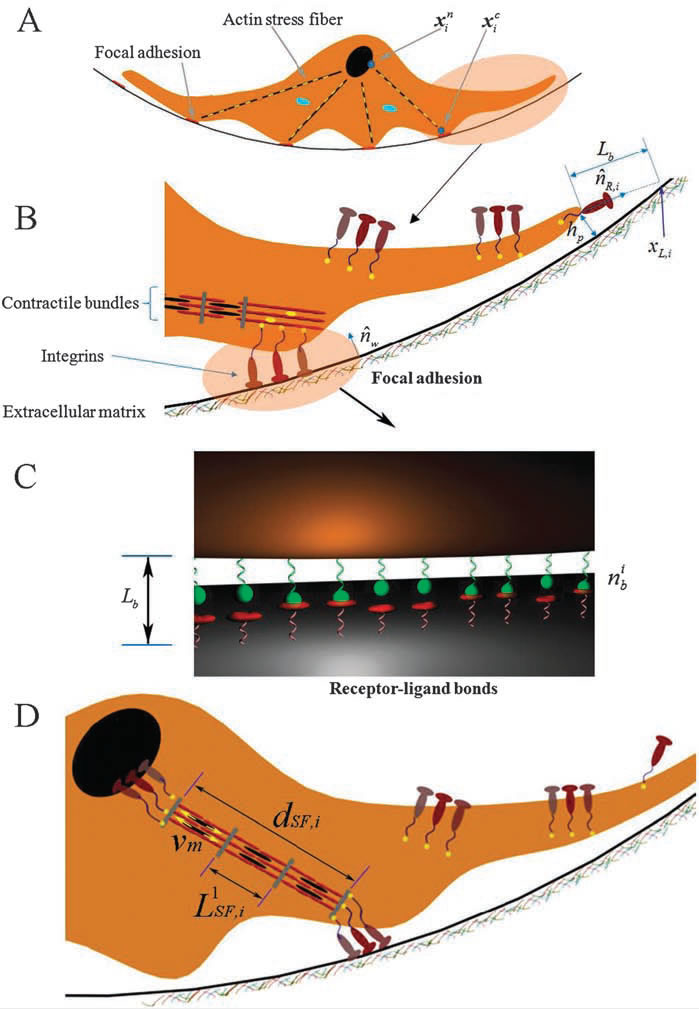
An integrative cell migration model incorporating focal adhesion (FA) dynamics, cytoskeleton and nucleus remodeling and actin motor activity is developed for predicting cell migration behaviors on 3-dimensional curved surfaces, such as cylindrical lumens in the 3-D extracellular matrix (ECM). The work is motivated by 3-D microfluidic migration experiments suggesting that the migration speed and direction may vary depending on the cross sectional shape of the lumen along which the cell migrates. In this paper, the mechanical structure of the cell is modeled as double elastic membranes of cell and nucleus. The two elastic membranes are connected by stress fibers, which are extended from focal adhesions on the cell surface to the nuclear membrane. The cell deforms and gains traction as transmembrane integrins distributed over the outer cell membrane bind to ligands on the ECM, form focal adhesions, and activate stress fibers. Probabilities at which integrin ligand–receptor bonds are formed as well as ruptures are affected by the surface geometry, resulting in diverse migration behaviors that depend on the curvature of the surface. Monte Carlo simulations of the integrative model reveal that (a) the cell migration speed is dependent on the cross sectional area of the lumen with a maximum speed at a particular diameter or width, (b) as the lumen diameter increases, the cell tends to spread and migrate around the circumference of the lumen, while it moves in the longitudinal direction as the lumen diameter narrows, (c) once the cell moves in one direction, it tends to stay migrating in the same direction despite the stochastic nature of migration. The relationship between the cell migration speed and the lumen width agrees with microfluidic experimental data for cancer cell migration.
|
|
|
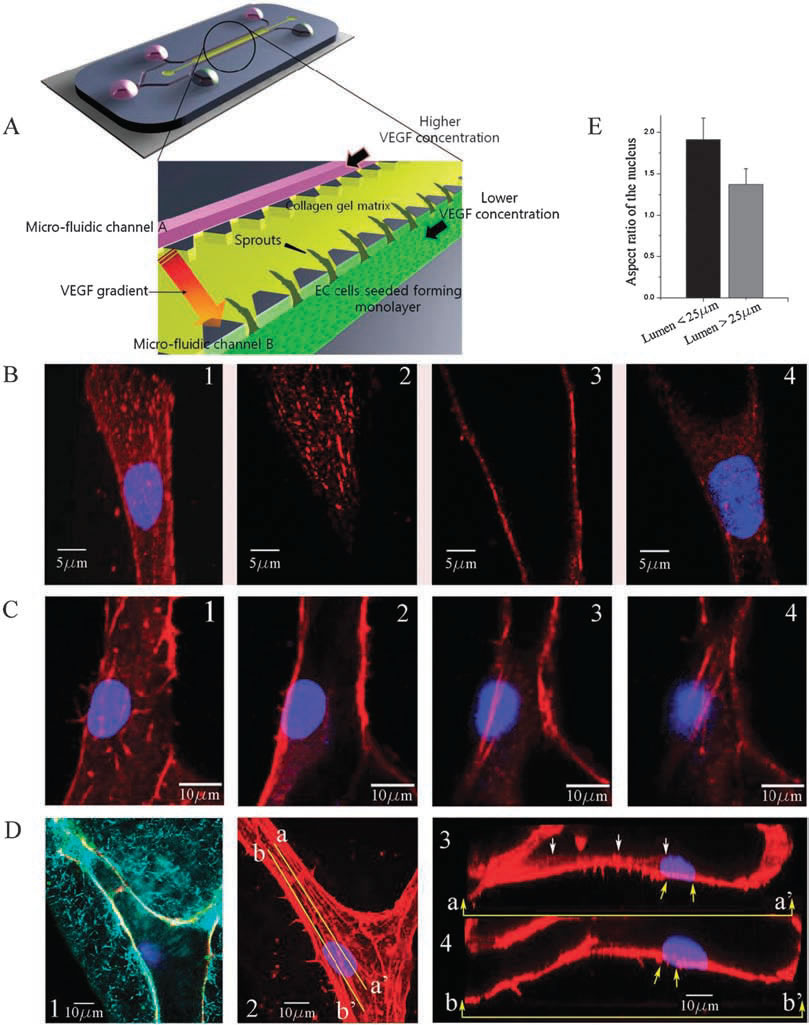
| Experimental observations of focal adhesion sites and actin stress fibers on the lumen in a 3-D collagen matrix: (A) 3-D microfluidic assay with hMVECs seeded on one side of the collagen gel. Higher concentration of VEGF is supplied to channel A and lower concentration of VEGF to channel B so that a gradient of VGEF is created across the gel. (B) Stalk cells migrating into the gel are observed; (1) collapsed confocal 3-D image (120X) showing a stalk cell migration along a narrow lumen, and slices at selected heights of (2) z=6.3 um (top), (3) 10.08 um (middle), and (4) 16.38 um (bottom); nucleus and focal adhesion sites are stained with Hoechst (blue) and vinculin (red), respectively. (C) Actin stress fibers in a larger lumen with a magnification of 120X; sectional slices showing stress fibers at selected heights; (1) z = 0 um, (B) 0.76 um, (3) 5.32 um and (4) 6.84 um; nucleus and actin stress fibers are stained with Hoechst (blue) and Rhodamine phalloidin (red), respectively. 6.84 um; nucleus and actin stress fibers are stained with Hoechst (blue) and Rhodamine phalloidin (red), respectively. (D(1)) A reflectance microscopy image showing the creation of a hole by a migrating tip cell in the 3-D collagen matrix; nucleus and vascular endothelial (VE)-cadherins are stained with Hoechst (blue) and anti-VE cadherin (green), (D(2)) collapsed confocal 3D image (120X); nucleus and actin stress fibers are stained with Hoechst (blue) and Rhodamine phalloidin (red), respectively, (D(3)) the longitudinal cross sectional view across line aa’ shown in D(2); short actin stress fibers are seen beneath the nucleus, (D(4)) the longitudinal cross sectional view across line bb’ shown in D(2); long stress fibers (yellow arrows) connected to the nucleus extend towards the leading edge of the migrating cell. (E) Quantification of the aspect ratio of the nucleus under two conditions when the cell migrates into the lumen with a diameter less than 25 mm and larger than 25 mm. Data are means +/- SD. In each case 35 cells were analysed. | |
|
|
|
Related publication:
Kim, M.-C., Kim, C., Wood, L., Neal, D., Kamm, R., and Asada, H., “Integrating Focal Adhesion Dynamics, Cytoskeleton Remodeling, and Actin Motor Activity for Predicting Cell Migration on 3D Curved Surfaces of Extracellular Matrix,” Integrative Biology, Vol. 4(11), pp. 1386-1397, September 2012.